Current issue
About the Journal
Scientific Council
Editorial Board
Regulatory and archival policy
Code of publishing ethics
Publisher
Information about the processing of personal data in relation to cookies and newsletter subscription
Archive
For Authors
For Reviewers
Contact
Reviewers
Annals reviewers in 2025
Annals reviewers in 2024
Annals reviewers in 2023
Annals reviewers in 2022
Annals reviewers in 2021
Annals reviewers in 2020
Annals reviewers in 2019
Annals reviewers in 2018
Annals reviewers in 2017
Annals reviewers in 2016
Annals reviewers in 2015
Annals reviewers in 2014
Annals reviewers in 2013
Annals reviewers in 2012
Links
Sklep Wydawnictwa SUM
Biblioteka Główna SUM
Śląski Uniwersytet Medyczny w Katowicach
Privacy policy
Accessibility statement
Reviewers
Annals reviewers in 2025
Annals reviewers in 2024
Annals reviewers in 2023
Annals reviewers in 2022
Annals reviewers in 2021
Annals reviewers in 2020
Annals reviewers in 2019
Annals reviewers in 2018
Annals reviewers in 2017
Annals reviewers in 2016
Annals reviewers in 2015
Annals reviewers in 2014
Annals reviewers in 2013
Annals reviewers in 2012
Individual gut microbiological signature in obese diabetic spouses – case report and literature review
1
Department of Internal Diseases Propaedeutics and Emergency Medicine, Faculty of Public Health in Bytom, Medical University of Silesia, Katowice, Poland
2
University of Applied Sciences in Nysa, Poland
Corresponding author
Magdalena Piłot
Katedra i Zakład Propedeutyki Chorób Wewnętrznych i Medycyny Ratunkowej, Wydział Zdrowia Publicznego w Bytomiu, Śląski Uniwersytet Medyczny w Katowicach, ul. Piekarska 18, 41-902 Bytom
Katedra i Zakład Propedeutyki Chorób Wewnętrznych i Medycyny Ratunkowej, Wydział Zdrowia Publicznego w Bytomiu, Śląski Uniwersytet Medyczny w Katowicach, ul. Piekarska 18, 41-902 Bytom
Ann. Acad. Med. Siles. 2024;78:31-40
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Due to the fact that the gut microbiome signature becomes more pronounced in type 2 diabetes, a better understanding of the role of microflora in diabetes (existing dysbiosis) provides new insight into the pathophysiology of this disorder. This study focused on the gut microbiome profiles of a married couple with type 2 diabetes and obesity living for last 35 years in a shared household in terms of their nutritional status, lifestyle and diabetes treatment methods. At the same time, an attempt was made to answer the question of which factors have the most significant impact on the intestinal microbiome.
Material and Methods:
Medical interviews of subjects, anthropometric measurements, body composition, 24-hour nutritional interviews, glycemic control, and stool samples were analyzed. The quantitative and qualitative examination of the fecal intestinal flora was performed by the next-generation sequencing method.
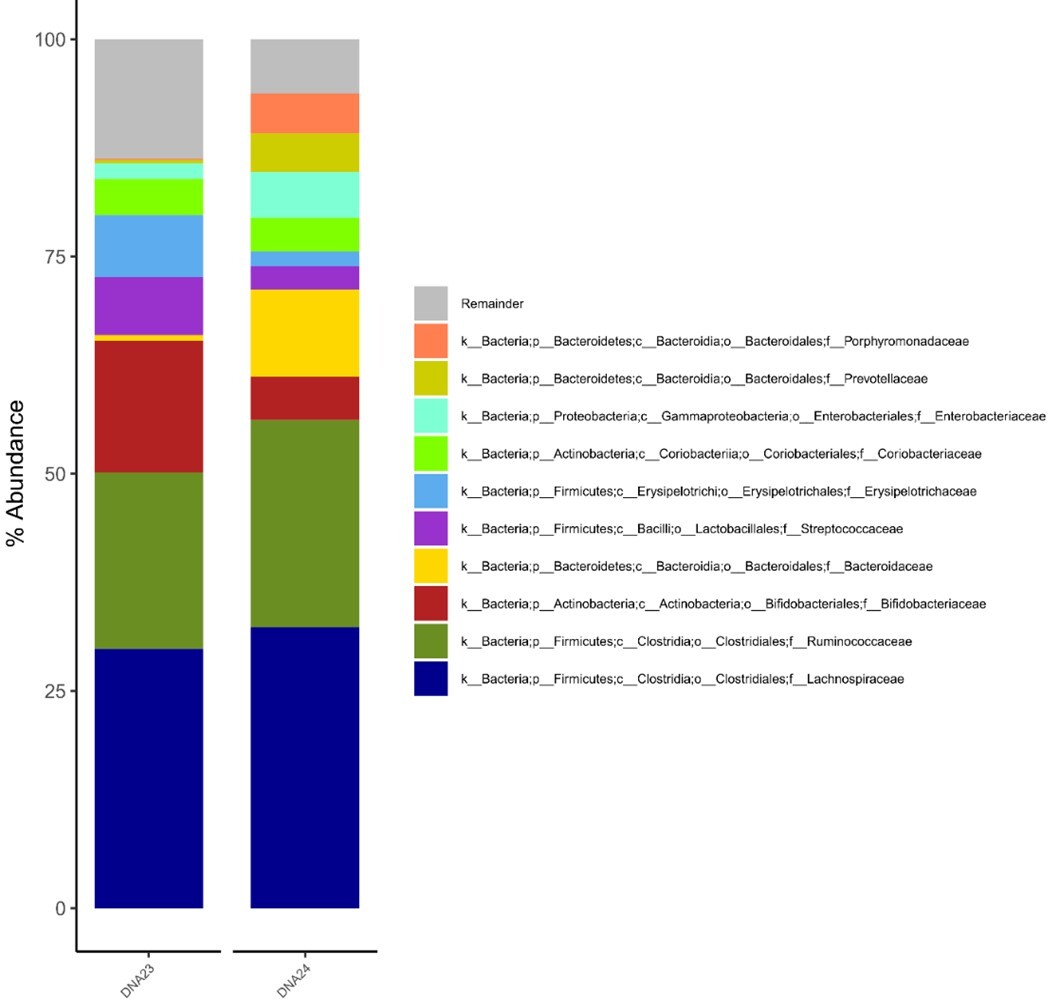
Results:
There were no significant differences in the study of the gut microbiome between the two subjects. The dominant bacterial phyla were Firmicutes and Actinobacteria, while Bacteroidetes and Proteobacteria shared smaller proportions, between 2 and 7%. Phylum Firmicutes was presented by the dominant Lachnospiraceae family (29–31%), Ruminococcaceae (16–19%), and Streptococcaceae (3–11%). The Actinobacteria phylum was proportionally less abundant and mainly represented by Bifidobacteriaceae (6–12%).
Conclusions:
May be the common living conditions have a significant influence on gut microbiota composition of diabetic spouses, despite differences in gender, comorbidities, diabetes therapy, diet and behaviors.
Due to the fact that the gut microbiome signature becomes more pronounced in type 2 diabetes, a better understanding of the role of microflora in diabetes (existing dysbiosis) provides new insight into the pathophysiology of this disorder. This study focused on the gut microbiome profiles of a married couple with type 2 diabetes and obesity living for last 35 years in a shared household in terms of their nutritional status, lifestyle and diabetes treatment methods. At the same time, an attempt was made to answer the question of which factors have the most significant impact on the intestinal microbiome.
Material and Methods:
Medical interviews of subjects, anthropometric measurements, body composition, 24-hour nutritional interviews, glycemic control, and stool samples were analyzed. The quantitative and qualitative examination of the fecal intestinal flora was performed by the next-generation sequencing method.
Results:
There were no significant differences in the study of the gut microbiome between the two subjects. The dominant bacterial phyla were Firmicutes and Actinobacteria, while Bacteroidetes and Proteobacteria shared smaller proportions, between 2 and 7%. Phylum Firmicutes was presented by the dominant Lachnospiraceae family (29–31%), Ruminococcaceae (16–19%), and Streptococcaceae (3–11%). The Actinobacteria phylum was proportionally less abundant and mainly represented by Bifidobacteriaceae (6–12%).
Conclusions:
May be the common living conditions have a significant influence on gut microbiota composition of diabetic spouses, despite differences in gender, comorbidities, diabetes therapy, diet and behaviors.
REFERENCES (48)
1.
Mithieux G. Gut microbiota and host metabolism: what relationship. Neuroendocrinology 2018; 106(4): 352–356, doi: 10.1159/000484526.
2.
Zhu B., Wang X., Li L. Human gut microbiome: the second genome of body. Protein Cell 2010; 1(8): 718–725, doi: 10.1007/s13238-010-0093-z.
3.
Li Q., Zhou J.M. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience 2016; 324: 131–139, doi: 10.1016/j.neuroscience.2016.03.013.
4.
Hattori M., Taylor T.D. The human intestinal microbiome: a new frontier of human biology. DNA Res. 2009; 16(1): 1–12, doi: 10.1093/dnares/dsn033.
5.
Giuffrè M., Campigotto M., Campisciano G., Comar M., Crocè L.S. A story of liver and gut microbes: how does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol. Gastrointest. Liver Physiol. 2020; 318(5): G889–G906, doi: 10.1152/ajpgi.00161.2019.
6.
Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013; 500(7464): 541–546, doi: 10.1038/nature12506.
7.
Giuffrè M., Moretti R., Campisciano G., da Silveira A.B.M., Monda V.M., Comar M., Di Bella S. et al. You talking to me? Says the enteric nerv-ous system (ENS) to the microbe. How intestinal microbes interact with the ENS. J. Clin. Med. 2020; 9(11): 3705, doi: 10.3390/jcm9113705.
8.
Clements S.J., Carding S.R. Diet, the intestinal microbiota, and immune health in aging. Crit. Rev. Food Sci. Nutr. 2018; 58(4): 651–661, doi: 10.1080/10408398.2016.1211086.
9.
Halmos T., Suba I. Physiological patterns of intestinal microbiota. The role of dysbacteriosis in obesity, insulin resistance, diabetes and metabolic syndrome. Orv. Hetil. 2016; 157(1): 13–22, doi: 10.1556/650.2015.30296.
10.
Delzenne N.M., Cani P.D., Everard A., Neyrinck A.M., Bindels L.B. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia 2015; 58(10): 2206–2217, doi: 10.1007/s00125-015-3712-7.
11.
Aydin Ö., Nieuwdorp M., Gerdes V. The gut microbiome as a target for the treatment of type 2 diabetes. Curr. Diab. Rep. 2018; 18(8): 55, doi: 10.1007/s11892-018-1020-6.
12.
Malczyk E., Dzięgielewska-Gęsiak S., Fatyga E., Ziółko E., Kokot T., Muc-Wierzgoń M. Body composition in healthy older persons: role of the ratio of extracellular/total body water. J. Biol. Regul. Homeost. Agents 2016; 30(3): 767–772.
13.
Szponar L., Wolnicka K., Rychlik E. Album of photographs of food products and dishes. National Food and Nutrition Institute. Warszawa 2000.
14.
Normy żywienia dla populacji Polski. M. Jarosz [ed.]. Instytut Żywności i Żywienia. Warszawa 2017.
15.
Jeżewska‐Zychowicz M., Gawęcki J., Wądołowska L., Czarnocińska J., Galiński G., Kołłajtis‐Dołowy A. et al. Kwestionariusz do badania poglądów i zwyczajów żywieniowych dla osób w wieku od 16 do 65 lat, wersja 1.2 – kwestionariusz do samodzielnego wypełnienia przez Respondenta. In: Kwestionariusz do badania poglądów i zwyczajów żywieniowych oraz procedura opracowania danych. J. Gawęcki [ed.]. Komitet Nauki o Żywieniu Człowieka Polskiej Akademii Nauk. Warszawa 2014, pp 21–33. Available at http://www.knozc.pan.pl/.
16.
Klindworth A., Preusse E., Schweer T., Peplies J., Quast C., Horn M. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acid. Res. 2013; 41(1): e1, doi: 10.1093/nar/gks808.
17.
Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014; 30(21): 3123–3124, doi: 10.1093/bioinformatics/btu494.
18.
Stackebrandt E. The Family Lachnospiraceae. In: The Prokaryotes: Firmicutes and Tenericutes. E. Rosenberg, E.F. DeLong, S. Lory, E. Stackebrandt, F. Thompson [ed.]. Springer. Berlin, Heidelberg 2014, pp 197–201, doi: 10.1007/978-3-642-30120-9_363.
19.
Dzięgielewska-Gęsiak S., Fatyga E., Piłot M., Wierzgoń A., Muc-Wierzgoń M. Are there differences in gut microbiome in patients with type 2 diabetes treated by metformin or metformin and insulin? Diabetes Metab. Syndr. Obes. 2022; 15: 3589–3599, doi: 10.2147/DMSO.S377856.
20.
Rinninella E., Raoul R., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A. et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019; 7(1): 14, doi: 10.3390/microorganisms7010014.
21.
Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444(7122): 1022–1023, doi: 10.1038/4441022a.
22.
Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. et al. Enterotypes of the human gut microbiome. Nature 2011; 473(7346): 174–180, doi: 10.1038/nature09944.
23.
Vacca M., Celano G., Calabrese F.M., Portincasa P., Gobbetti M., De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020; 8(4): 573, doi: 10.3390/microorganisms8040573.
24.
Zeng H., Ishaq S.L., Zhao F.Q., Wright A.D.G. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016; 35: 30–36, doi: 10.1016/j.jnutbio.2016.05.015.
25.
Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013; 7(4): 880–884, doi: 10.1038/ismej.2012.153.
26.
Takagi T., Naito Y., Kashiwagi S., Uchiyama K., Mizushima K., Kamada K. et al. Changes in the gut microbiota are associated with hypertension, hyperlipidemia, and type 2 diabetes mellitus in Japanese subjects. Nutrients 2020; 12(10): 2996, doi: 10.3390/nu12102996.
27.
Salonen A., Lahti L., Salojärvi J., Holtrop G., Korpela K., Duncan S.H. et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014; 8(11): 2218–2230, doi: 10.1038/ismej.2014.63.
28.
Hashimoto Y., Hamaguchi M., Kaji A., Sakai R., Osaka T., Inoue R. et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J. Diabetes Investig. 2020; 11(6): 1623–1634, doi: 10.1111/jdi.13293.
29.
Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490(7418): 55–60, doi: 10.1038/nature11450.
30.
Adachi K., Sugiyama T., Yamaguchi Y., Tamura Y., Izawa S., Hijikata Y. et al. Gut microbiota disorders cause type 2 diabetes mellitus and homeostatic disturbances in gut-related metabolism in Japanese subjects. J. Clin. Biochem. Nutr 2019; 64(3): 231–238, doi: 10.3164/jcbn.18-101.
31.
Santos-Marcos J.A., Rangel-Zuñiga O.A., Jimenez-Lucena R., Quintana-Navarro G.M., Garcia-Carpintero S., Malagon M.M. et al. Influence of gender and menopausal status on gut microbiota. Maturitas 2018; 116: 43–53, doi: 10.1016/j.maturitas.2018.07.008.
32.
Larsen N., Vogensen F.K., van den Berg F.W.J., Nielsen D.S., Andreasen A.S., Pedersen B.K. et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010; 5(2): e9085, doi: 10.1371/journal.pone.0009085.
33.
de la Cuesta-Zuluaga J., Mueller N.T., Corrales-Agudelo V., Velásquez-Mejía E.P., Carmona J.A., Abad J.M. et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 2017; 40(1): 54–62, doi: 10.2337/dc16-1324.
34.
Elbere I., Kalnina I., Silamikelis I., Konrade I., Zaharenko L., Sekace K. et al. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS One 2018; 13(9): e0204317, doi: 10.1371/journal.pone.0204317.
35.
Wang H., Tang W., Zhang P., Zhang Z., Jielei H., Zhu D. et al. Modulation of gut microbiota contributes to effects of intensive insulin therapy on intestinal morphological alteration in high-fat-diet-treated mice. Acta Diabetol. 2020; 57(4): 455–467, doi: 10.1007/s00592-019-01436-0.
36.
Savin Z., Kivity S., Yonath H., Yehuda S. Smoking and the intestinal microbiome. Arch. Microbiol. 2018; 200(5): 677–684, doi: 10.1007/s00203-018-1506-2.
37.
Sublette M.G., Cross T.L., Korcarz C.E., Hansen K.M., Murga-Garrido S.M., Hazen S.L. et al. Effects of smoking and smoking cessation on the intestinal microbiota. J. Clin. Med. 2020; 9(9): 2963, doi: 10.3390/jcm9092963.
38.
Lee S.H., Yun Y., Kim S.J., Lee E.J., Chang Y., Ryu S. et al. Association between cigarette smoking status and composition of gut microbiota: population-based cross-sectional study. J. Clin. Med. 2018; 7(9): 282, doi: 10.3390/jcm7090282.
39.
David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505(7484): 559–563, doi: 10.1038/nature12820.
40.
Scott K.P., Gratz S.W., Sheridan P.O., Flint H.J., Duncan S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013; 69(1): 52–60, doi: 10.1016/j.phrs.2012.10.020.
41.
Magnúsdóttir S., Ravcheev D., de Crécy-Lagard V., Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015; 6: 148, doi: 10.3389/fgene.2015.00148.
42.
Ojo O., Feng Q.Q., Ojo O.O., Wang X.H. The role of dietary fibre in modulating gut microbiota dysbiosis in patients with type 2 diabetes: a systematic review and meta-analysis of randomised controlled trials. Nutrients 2020; 12(11): 3239, doi: 10.3390/nu12113239.
43.
Makki K., Deehan E.C., Walter J., Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018; 23(6): 705–715, doi: 10.1016/j.chom.2018.05.012.
44.
Schwiertz A., Taras D., Schäfer K., Beijer S., Bos N.A., Donus C. et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010; 18(1): 190–195, doi: 10.1038/oby.2009.167.
45.
Herd P., Schaeffer N.C., DiLoreto K., Jacques K., Stevenson J., Rey F. et al. The influence of social conditions across the life course on the human gut microbiota: a pilot project with the Wisconsin Longitudinal Study. J. Gerontol. B. Psychol. Sci. Soc. Sci. 2017; 73(1): 124–133, doi: 10.1093/geronb/gbx029.
46.
Dic-McFarland K.A., Tang Z.Z., Kemis J.H., Kerby R.L., Chen G., Palloni A. et al. Close social relationships correlate with human gut microbiota composition. Sci. Rep. 2019; 24; 9(1): 703, doi: 10.1038/s41598-018-37298-9.
47.
Bleau C., Karelis A.D., St-Pierre D.H., Lamontagne L. Crosstalk between intestinal microbiota, adipose tissue and skeletal muscle as an early event in systemic low-grade inflammation and the development of obesity and diabetes. Diabetes Metab. Res. Rev. 2015; 31(6): 545–561, https://doi.org/10.1002/dmrr.2....
48.
Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334(6052): 105–108, doi: 10.1126/science.1208344.
CITATIONS (1):
1.
Gut Microbiota and Metabolic Dysregulation in Elderly Diabetic Patients: Is There a Gender-Specific Effect
Magdalena Piłot, Sylwia Dzięgielewska-Gęsiak, Katarzyna Weronika Walkiewicz, Martyna Bednarczyk, Dariusz Waniczek, Małgorzata Muc-Wierzgoń
Journal of Clinical Medicine
Magdalena Piłot, Sylwia Dzięgielewska-Gęsiak, Katarzyna Weronika Walkiewicz, Martyna Bednarczyk, Dariusz Waniczek, Małgorzata Muc-Wierzgoń
Journal of Clinical Medicine
Share
RELATED ARTICLE
The Medical University of Silesia in Katowice, as the Operator of the annales.sum.edu.pl website, processes personal data collected when visiting the website. The function of obtaining information about Users and their behavior is carried out by voluntarily entered information in forms, saving cookies in end devices, as well as by collecting web server logs, which are in the possession of the website Operator. Data, including cookies, are used to provide services in accordance with the Privacy policy.
You can consent to the processing of data for these purposes, refuse consent or access more detailed information.
You can consent to the processing of data for these purposes, refuse consent or access more detailed information.